Extract conditional posterior means and standard deviations, marginal posterior means and standard deviations, posterior probabilities, and marginal inclusions probabilities under Bayesian Model Averaging from an object of class 'bas'
Arguments
- object
object of class 'bas' created by BAS
- n.models
Number of top models to report in the printed summary, for coef the default is to use all models. To extract summaries for the Highest Probability Model, use n.models=1 or estimator="HPM".
- estimator
return summaries for a selected model, rather than using BMA. Options are 'HPM' (highest posterior probability model) ,'MPM' (median probability model), and 'BMA'
- ...
other optional arguments
- x
object of class 'coef.bas' to print
- digits
number of significant digits to print
Value
coefficients returns an object of class coef.bas with the
following:
- conditionalmeans
a matrix with conditional posterior means for each model
- conditionalsd
standard deviations for each model
- postmean
marginal posterior means of each regression coefficient using BMA
- postsd
marginal posterior standard deviations using BMA
- postne0
vector of posterior inclusion probabilities, marginal probability that a coefficient is non-zero
Details
Calculates posterior means and (approximate) standard deviations of the regression coefficients under Bayesian Model averaging using g-priors and mixtures of g-priors. Print returns overall summaries. For fully Bayesian methods that place a prior on g, the posterior standard deviations do not take into account full uncertainty regarding g. Will be updated in future releases.
Note
With highly correlated variables, marginal summaries may not be
representative of the joint distribution. Use plot.coef.bas to
view distributions. The value reported for the intercept is
under the centered parameterization. Under the Gaussian error
model it will be centered at the sample mean of Y.
References
Liang, F., Paulo, R., Molina, G., Clyde, M. and Berger, J.O.
(2005) Mixtures of g-priors for Bayesian Variable Selection. Journal of the
American Statistical Association. 103:410-423.
doi:10.1198/016214507000001337
Author
Merlise Clyde clyde@duke.edu
Examples
data("Hald")
hald.gprior = bas.lm(Y~ ., data=Hald, n.models=2^4, alpha=13,
prior="ZS-null", initprobs="Uniform", update=10)
coef.hald.gprior = coefficients(hald.gprior)
coef.hald.gprior
#>
#> Marginal Posterior Summaries of Coefficients:
#>
#> Using BMA
#>
#> Based on the top 16 models
#> post mean post SD post p(B != 0)
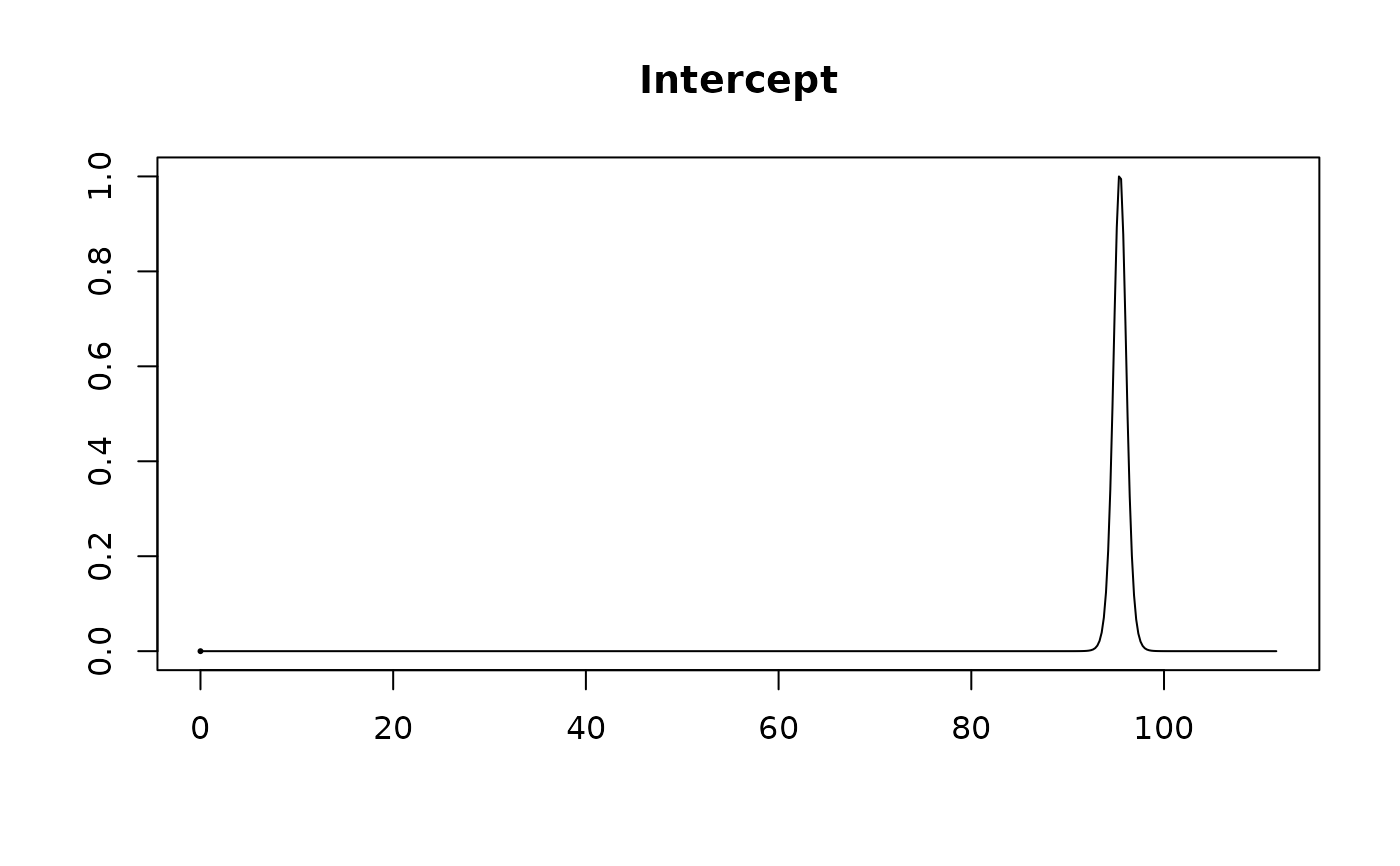
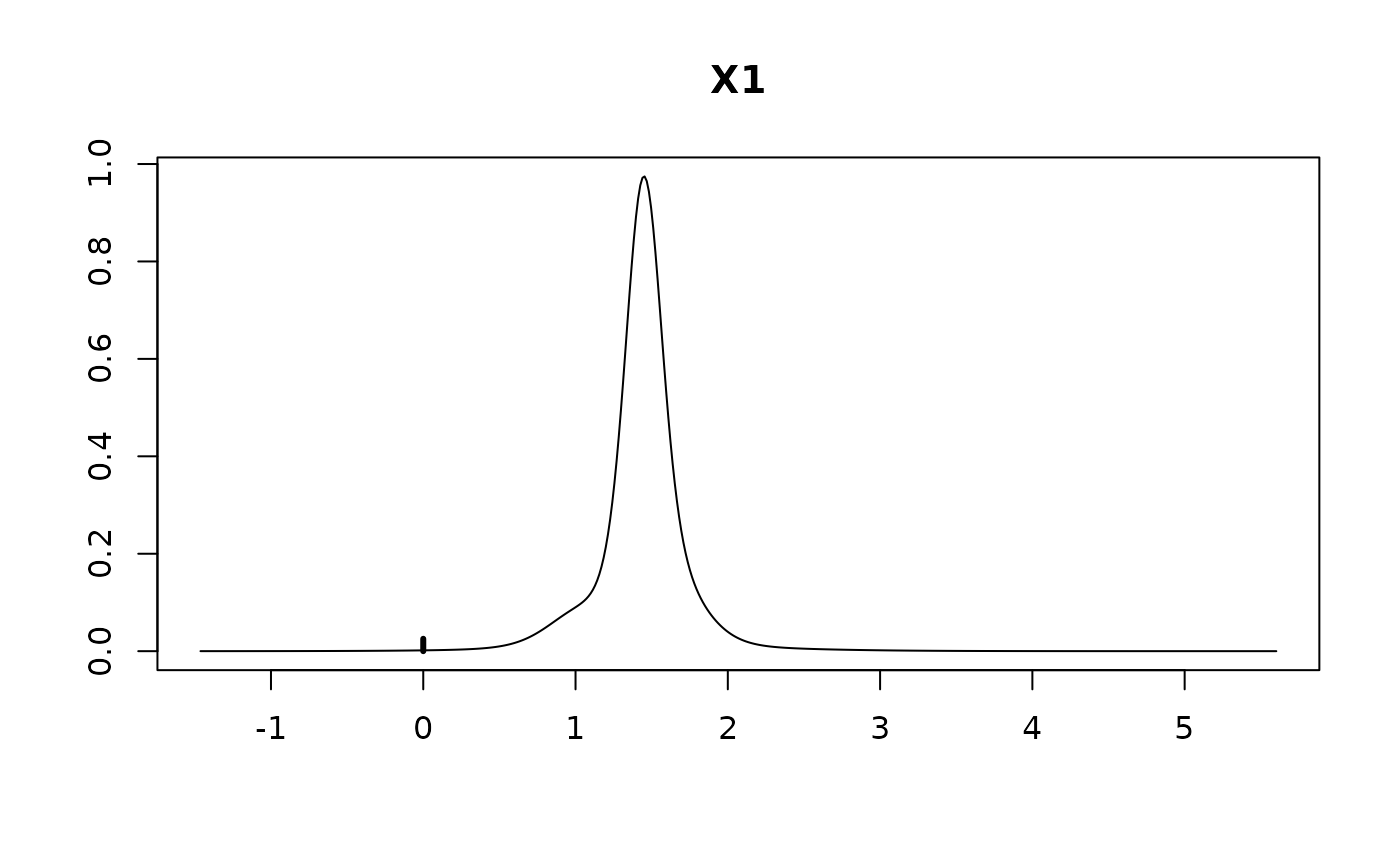
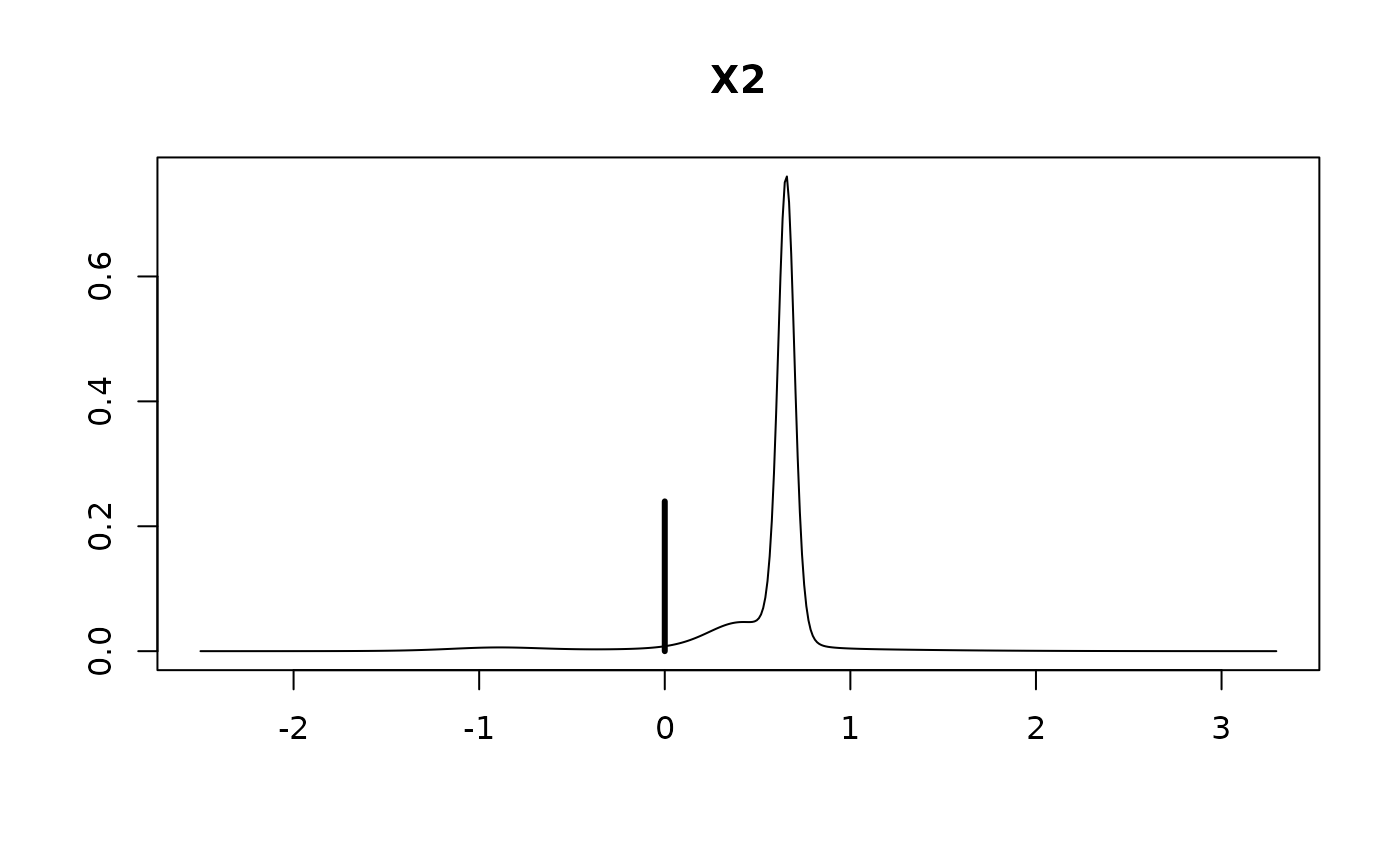
#> Intercept 95.42308 0.67885 1.00000
#> X1 1.40116 0.35351 0.97454
#> X2 0.42326 0.38407 0.76017
#> X3 -0.03997 0.33398 0.30660
#> X4 -0.22077 0.36931 0.44354
plot(coef.hald.gprior)
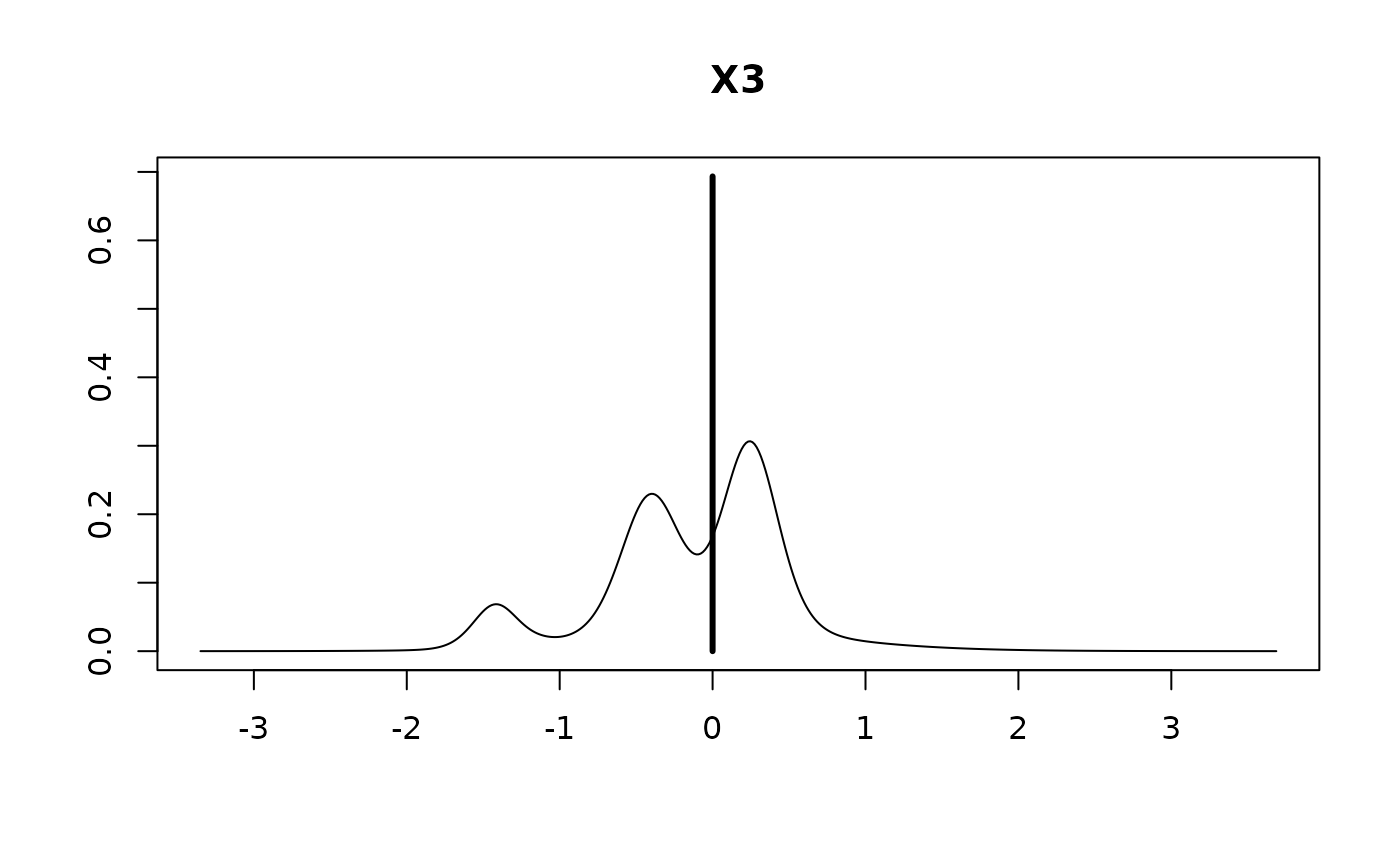
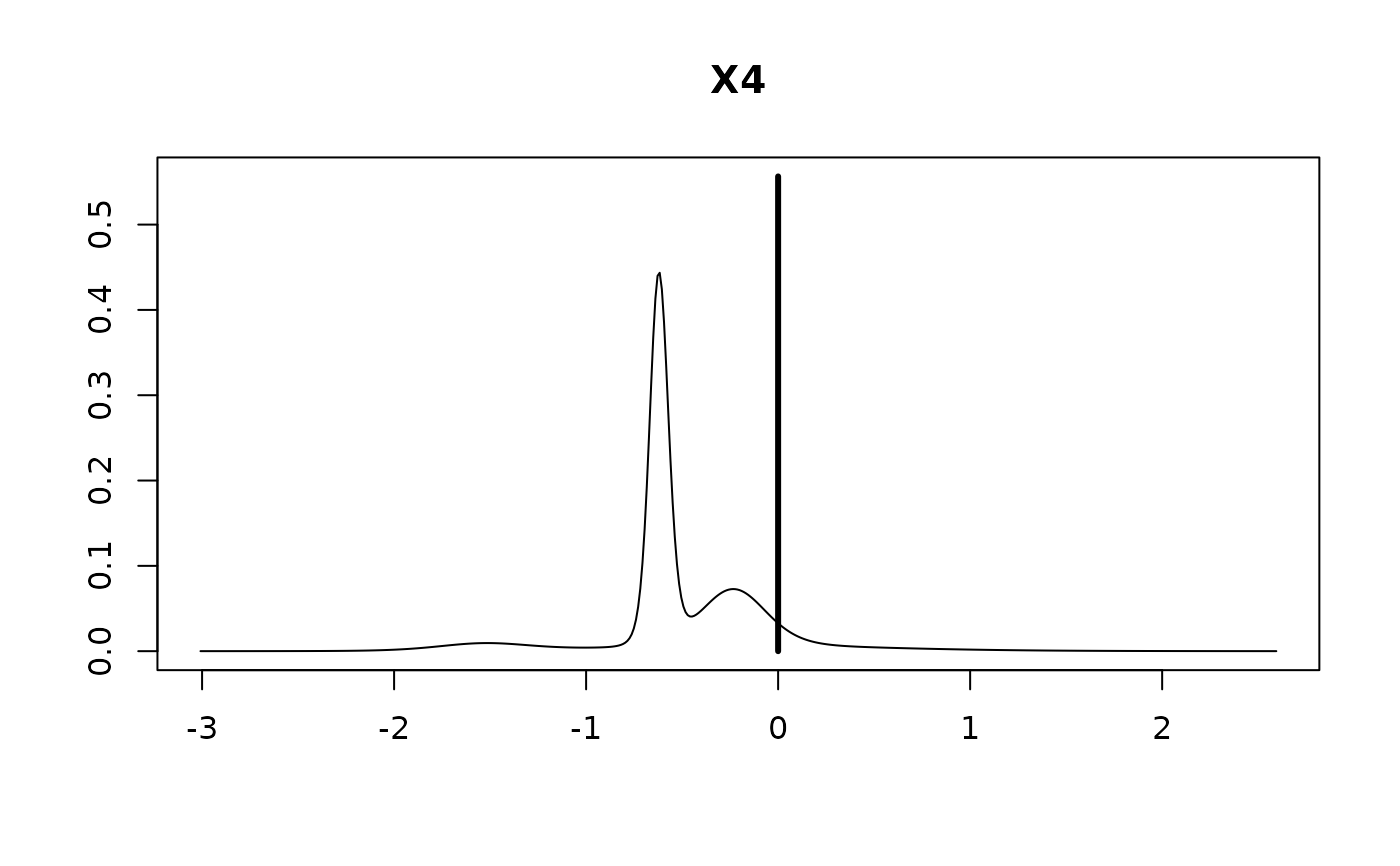 confint(coef.hald.gprior)
#> 2.5% 97.5% beta
#> Intercept 93.9711411 96.8741681 95.42307692
#> X1 0.6422478 2.0636407 1.40116123
#> X2 -0.0271815 0.8707052 0.42325794
#> X3 -0.7729096 0.5982604 -0.03997087
#> X4 -0.7461698 0.1970682 -0.22076600
#> attr(,"Probability")
#> [1] 0.95
#> attr(,"class")
#> [1] "confint.bas"
#Estimation under Median Probability Model
coef.hald.gprior = coefficients(hald.gprior, estimator="MPM")
coef.hald.gprior
#>
#> Marginal Posterior Summaries of Coefficients:
#>
#> Using MPM
#>
#> Based on the top 1 models
#> post mean post SD post p(B != 0)
#> Intercept 95.42308 0.66740 1.00000
#> X1 1.45542 0.12077 1.00000
#> X2 0.65644 0.04565 1.00000
#> X3 0.00000 0.00000 0.00000
#> X4 0.00000 0.00000 0.00000
plot(coef.hald.gprior)
confint(coef.hald.gprior)
#> 2.5% 97.5% beta
#> Intercept 93.9711411 96.8741681 95.42307692
#> X1 0.6422478 2.0636407 1.40116123
#> X2 -0.0271815 0.8707052 0.42325794
#> X3 -0.7729096 0.5982604 -0.03997087
#> X4 -0.7461698 0.1970682 -0.22076600
#> attr(,"Probability")
#> [1] 0.95
#> attr(,"class")
#> [1] "confint.bas"
#Estimation under Median Probability Model
coef.hald.gprior = coefficients(hald.gprior, estimator="MPM")
coef.hald.gprior
#>
#> Marginal Posterior Summaries of Coefficients:
#>
#> Using MPM
#>
#> Based on the top 1 models
#> post mean post SD post p(B != 0)
#> Intercept 95.42308 0.66740 1.00000
#> X1 1.45542 0.12077 1.00000
#> X2 0.65644 0.04565 1.00000
#> X3 0.00000 0.00000 0.00000
#> X4 0.00000 0.00000 0.00000
plot(coef.hald.gprior)
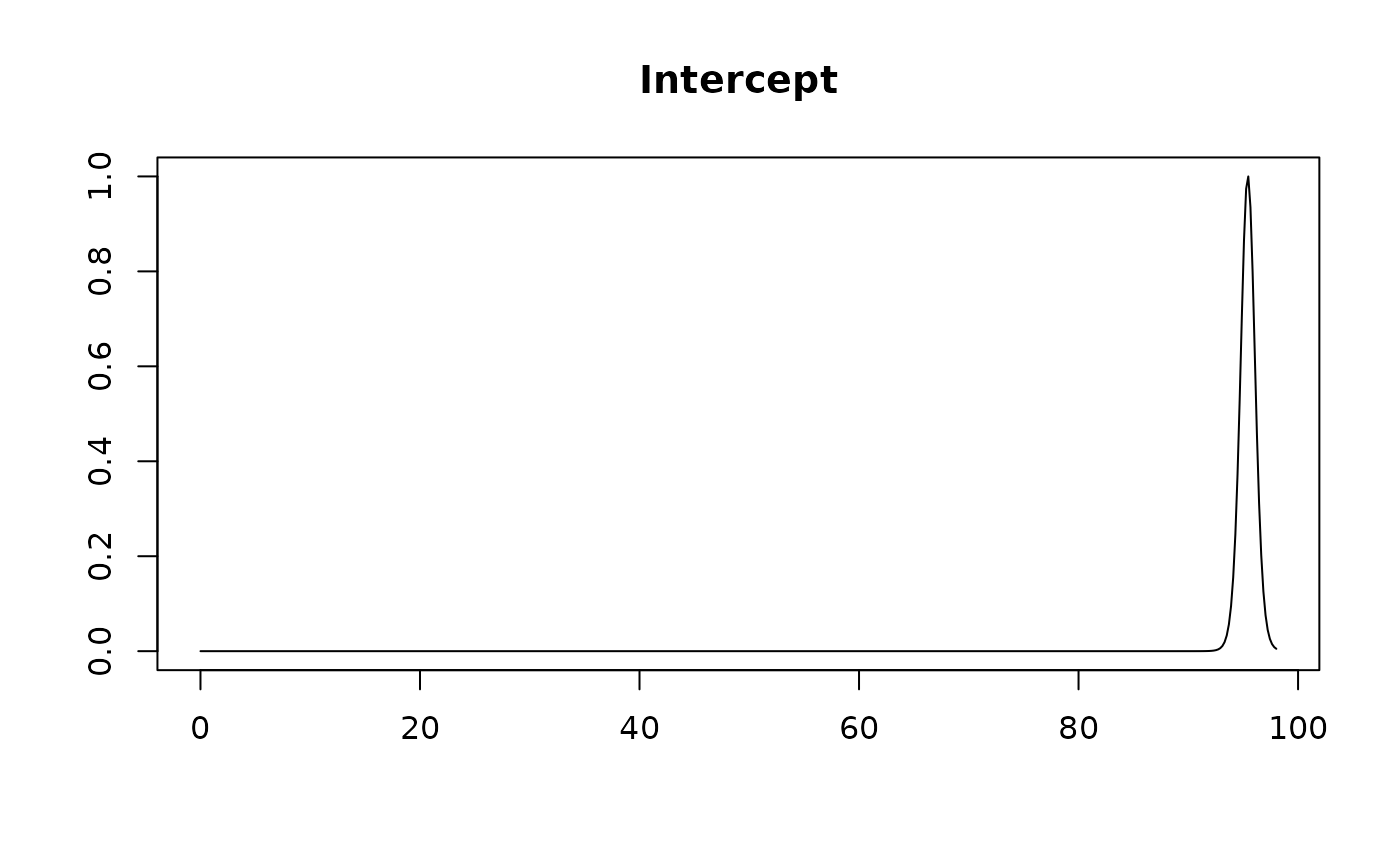
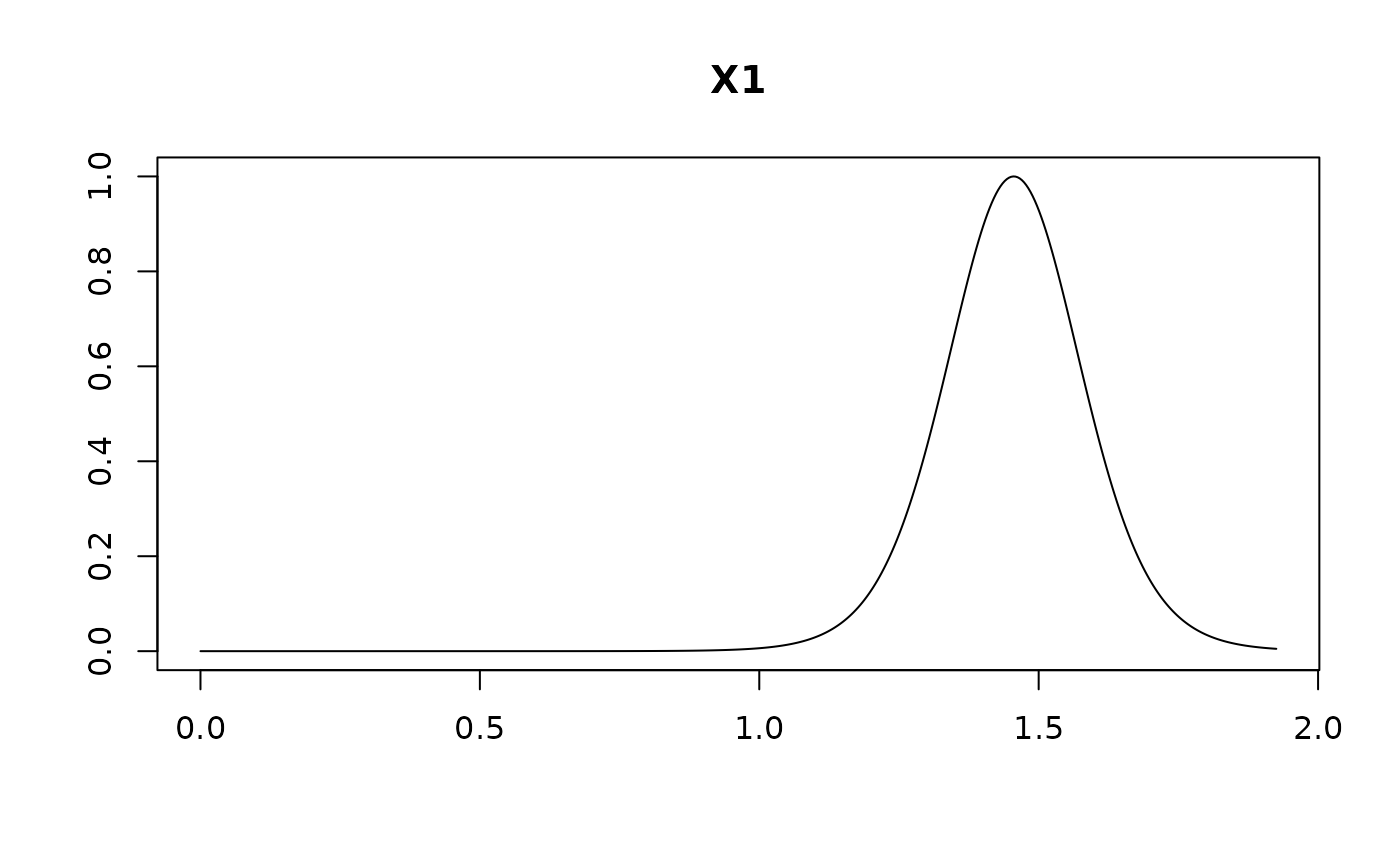
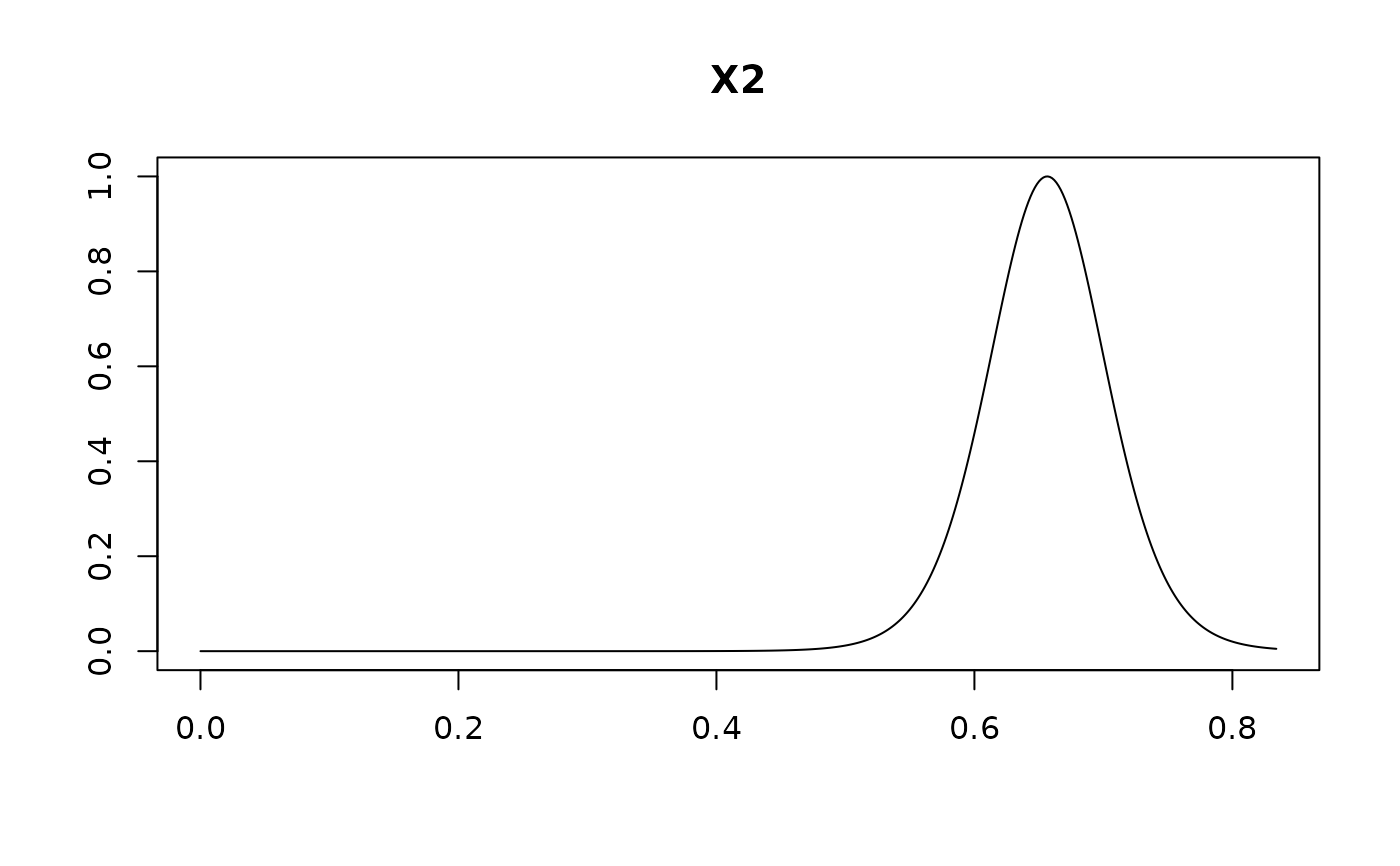
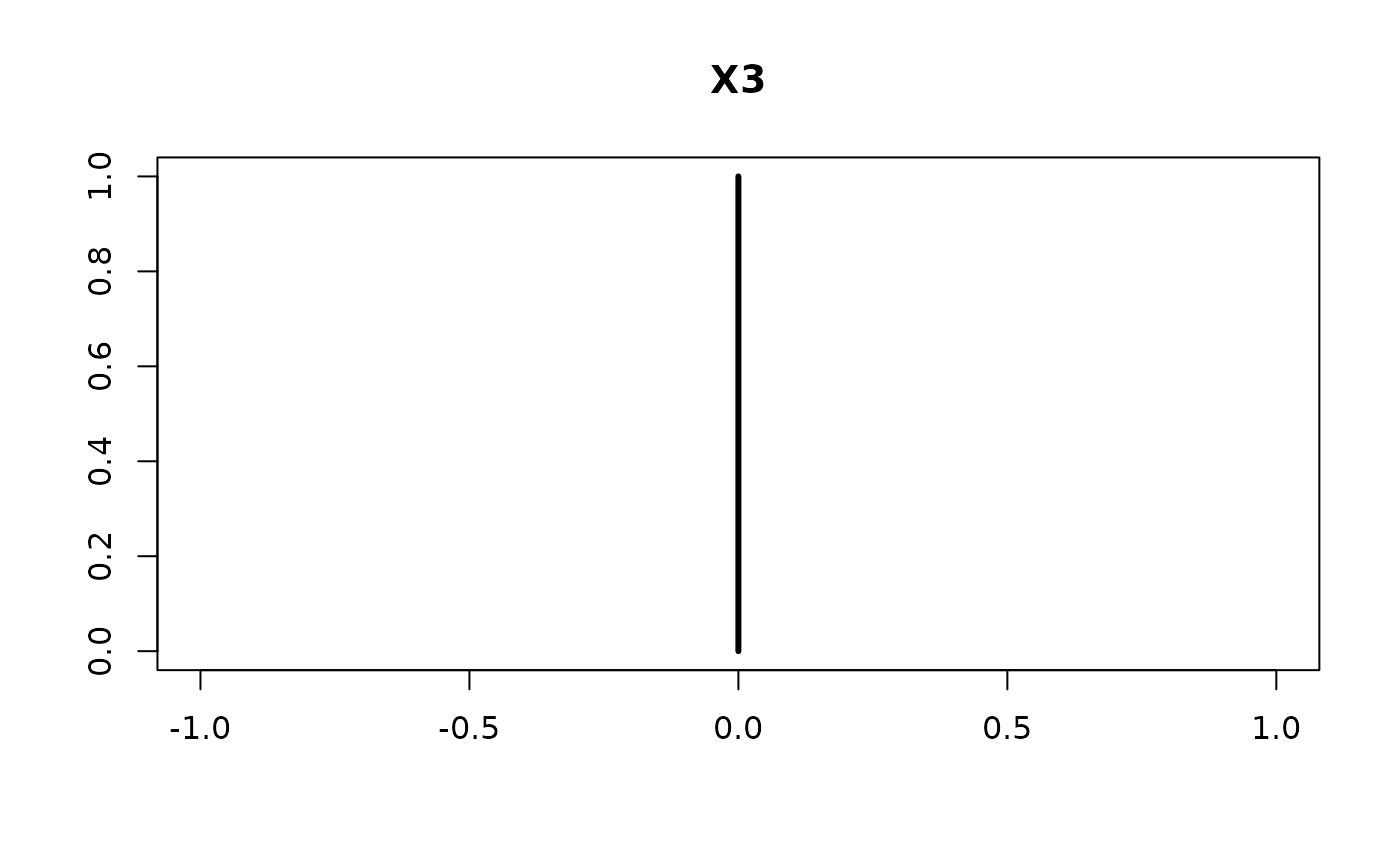
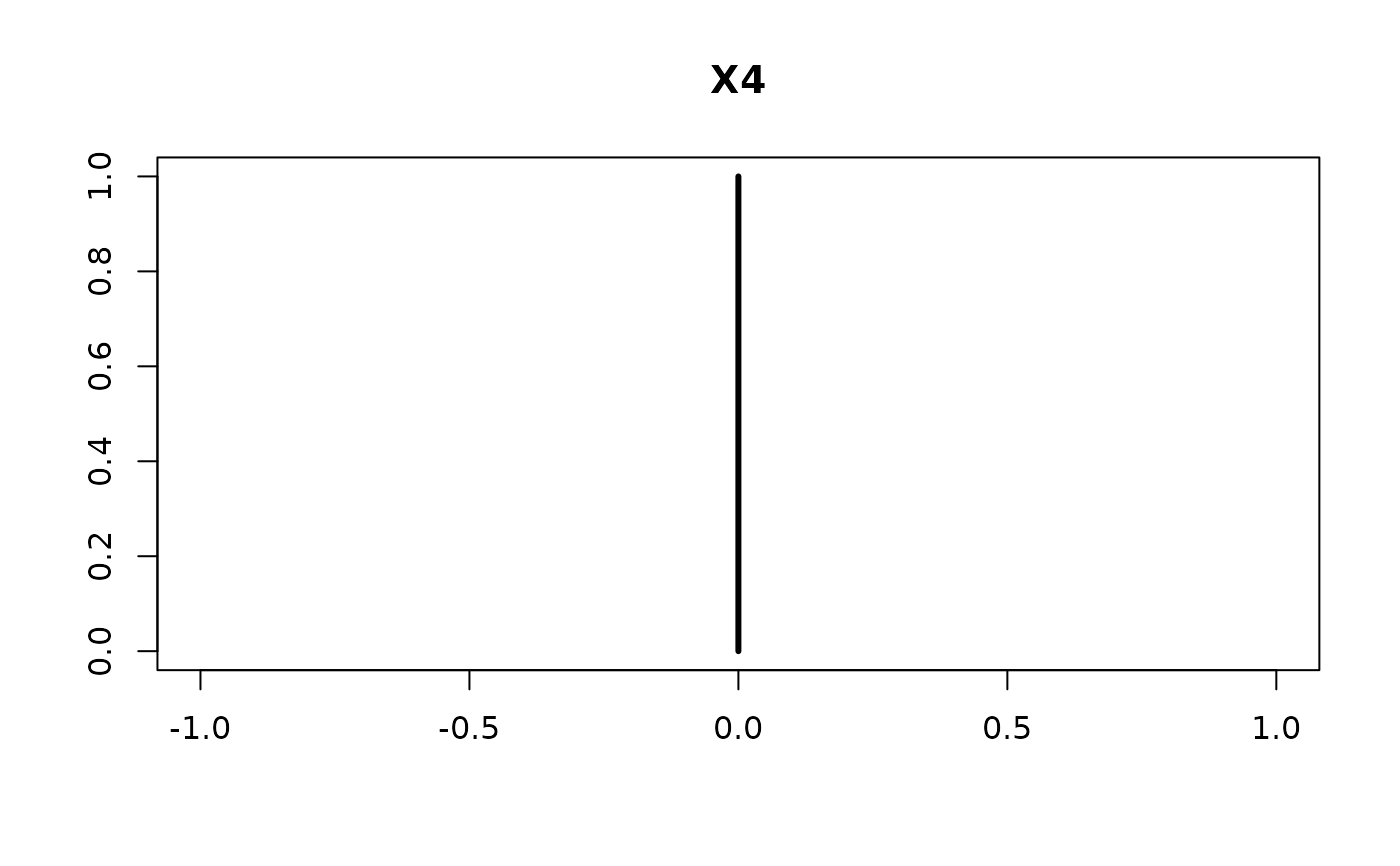 plot(confint(coef.hald.gprior))
plot(confint(coef.hald.gprior))
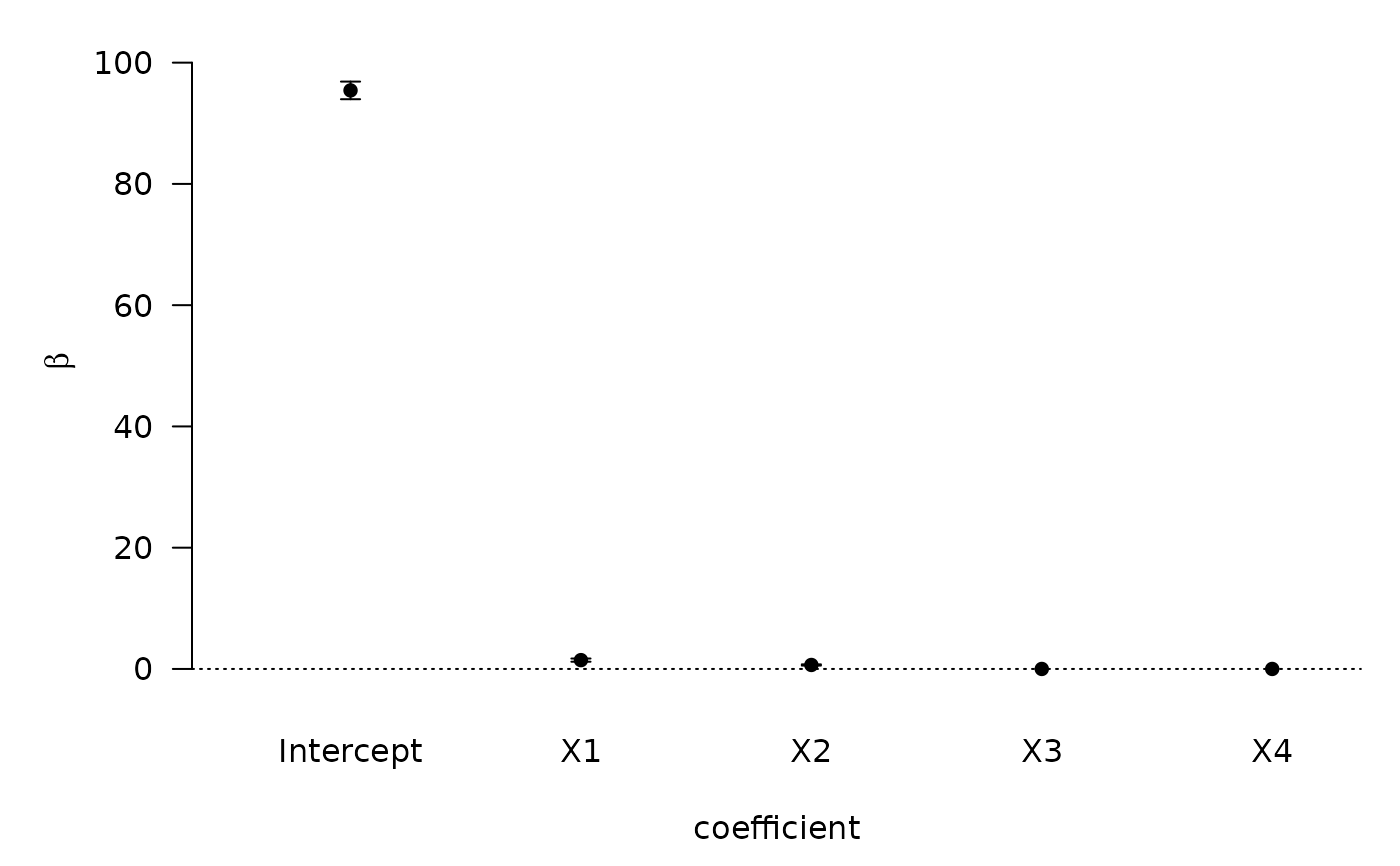 #> NULL
coef.hald.gprior = coefficients(hald.gprior, estimator="HPM")
coef.hald.gprior
#>
#> Marginal Posterior Summaries of Coefficients:
#>
#> Using HPM
#>
#> Based on the top 1 models
#> post mean post SD post p(B != 0)
#> Intercept 95.42308 0.66740 1.00000
#> X1 1.45542 0.12077 0.97454
#> X2 0.65644 0.04565 0.76017
#> X3 0.00000 0.00000 0.30660
#> X4 0.00000 0.00000 0.44354
plot(coef.hald.gprior)
#> NULL
coef.hald.gprior = coefficients(hald.gprior, estimator="HPM")
coef.hald.gprior
#>
#> Marginal Posterior Summaries of Coefficients:
#>
#> Using HPM
#>
#> Based on the top 1 models
#> post mean post SD post p(B != 0)
#> Intercept 95.42308 0.66740 1.00000
#> X1 1.45542 0.12077 0.97454
#> X2 0.65644 0.04565 0.76017
#> X3 0.00000 0.00000 0.30660
#> X4 0.00000 0.00000 0.44354
plot(coef.hald.gprior)
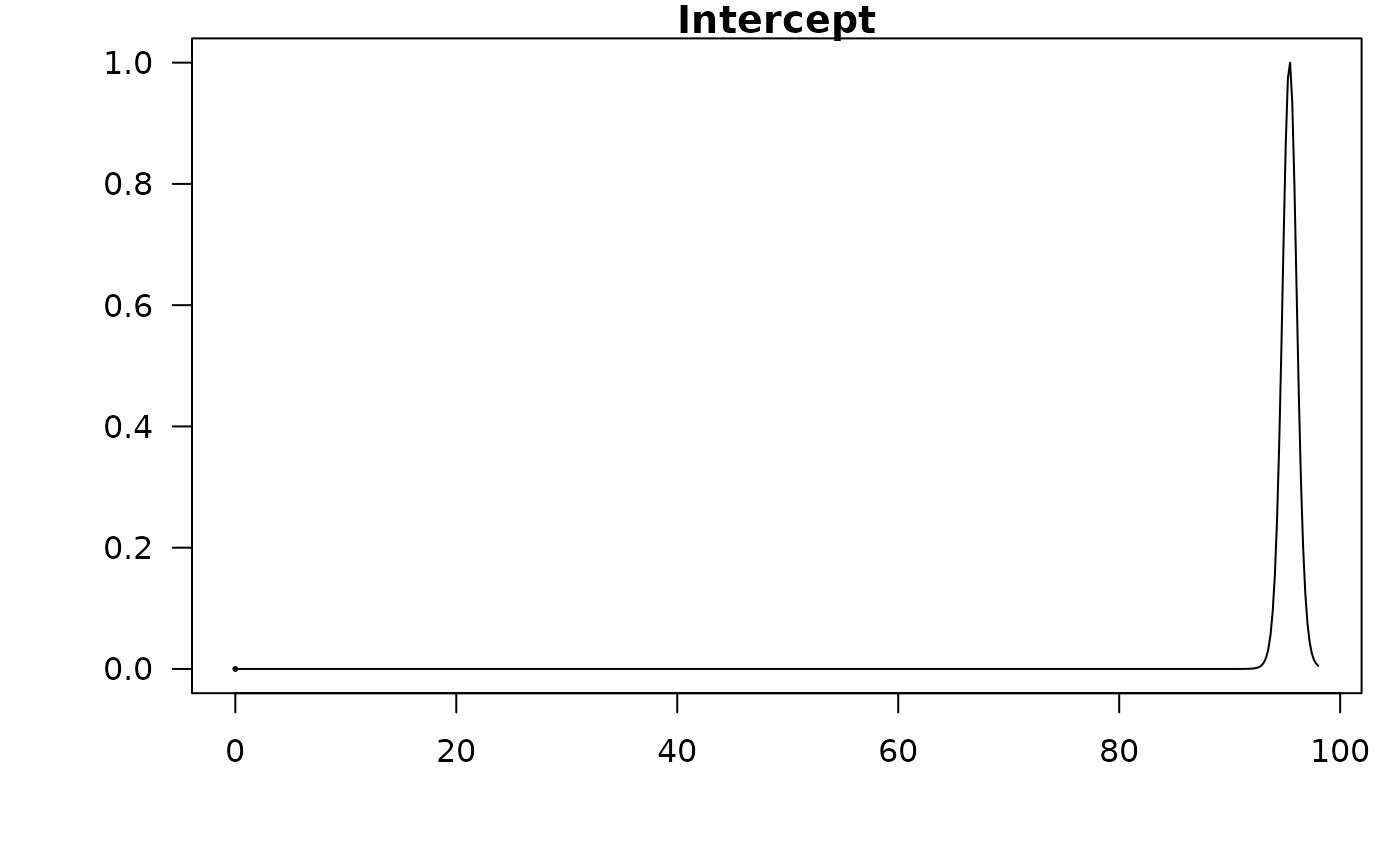
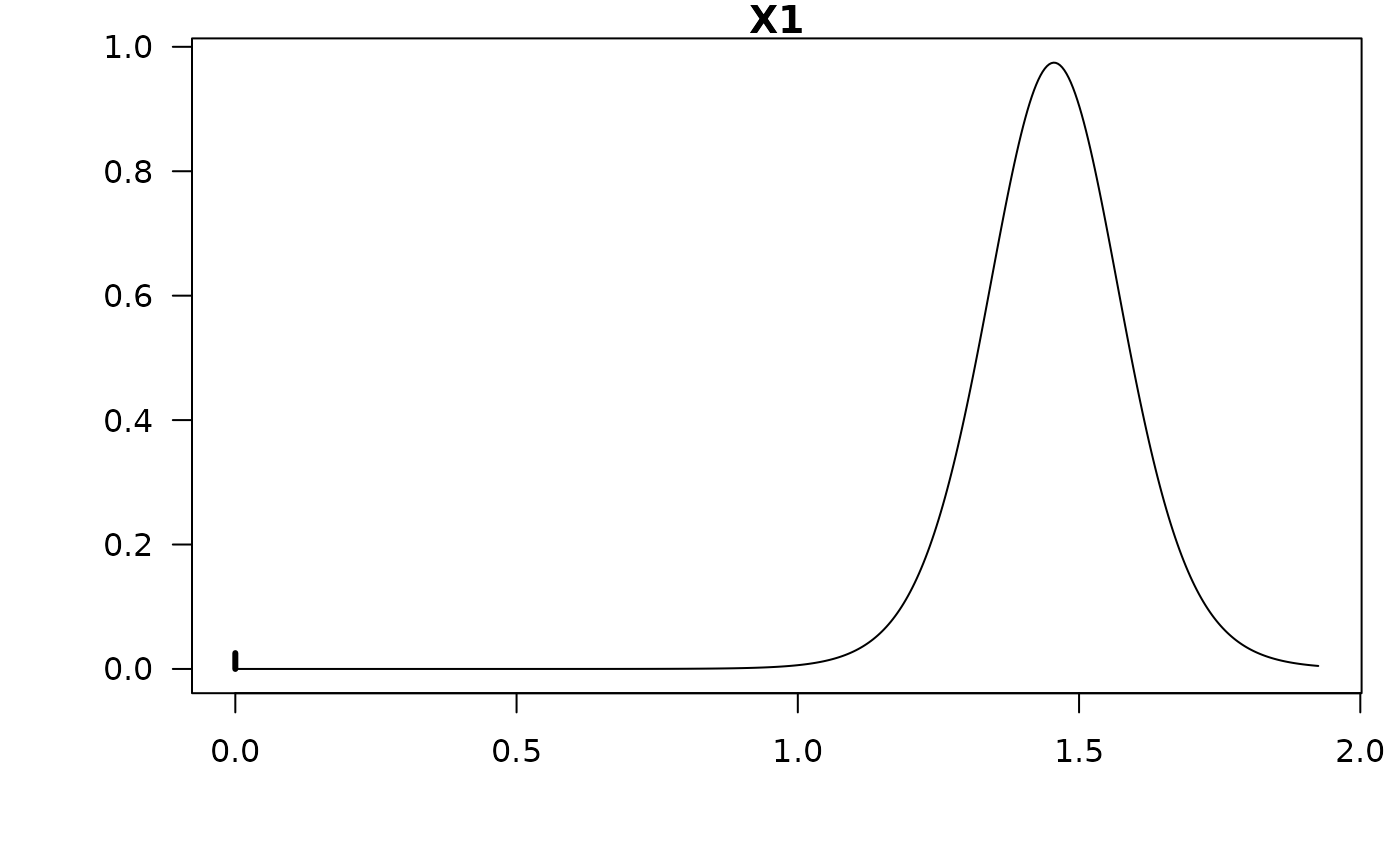
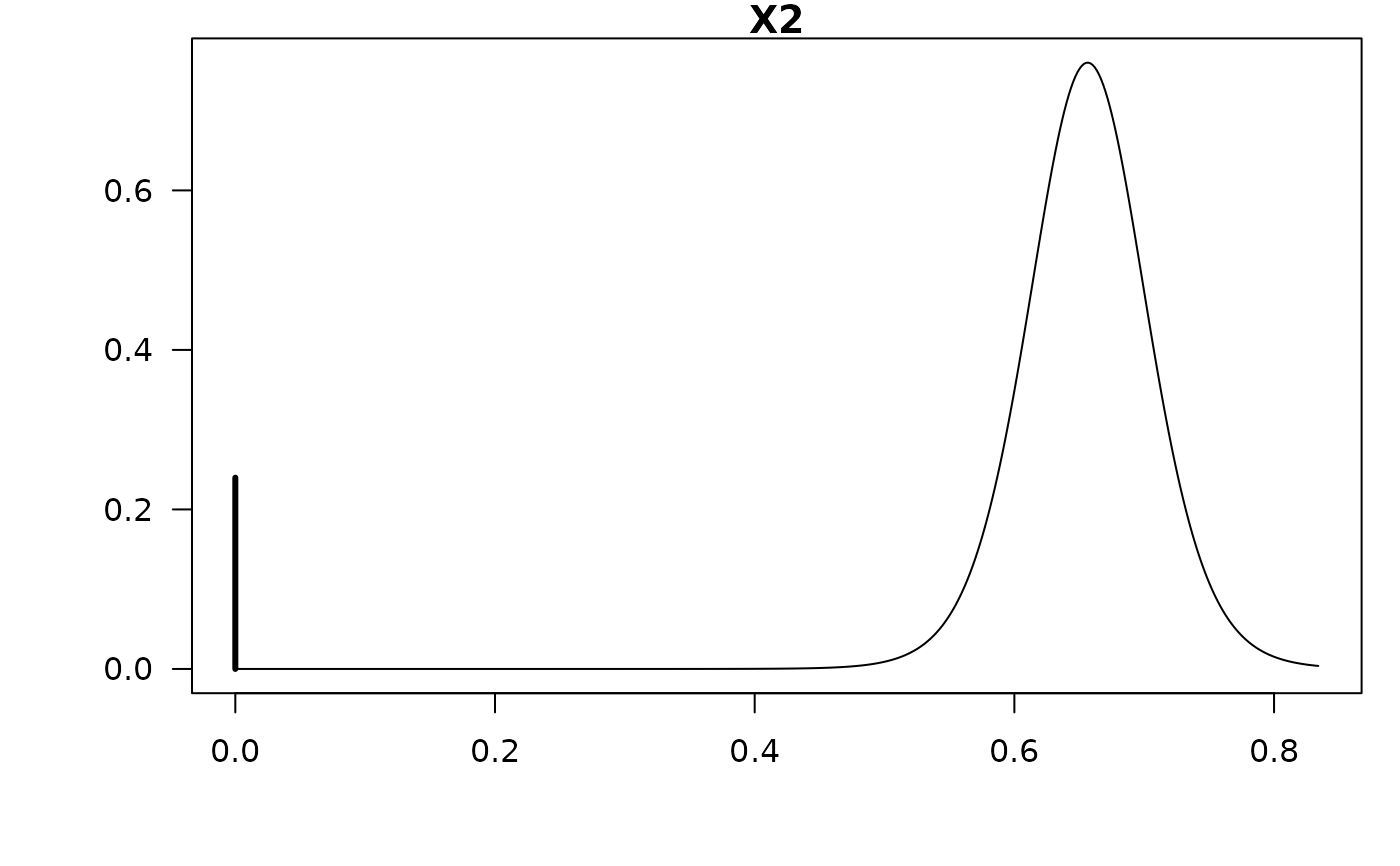
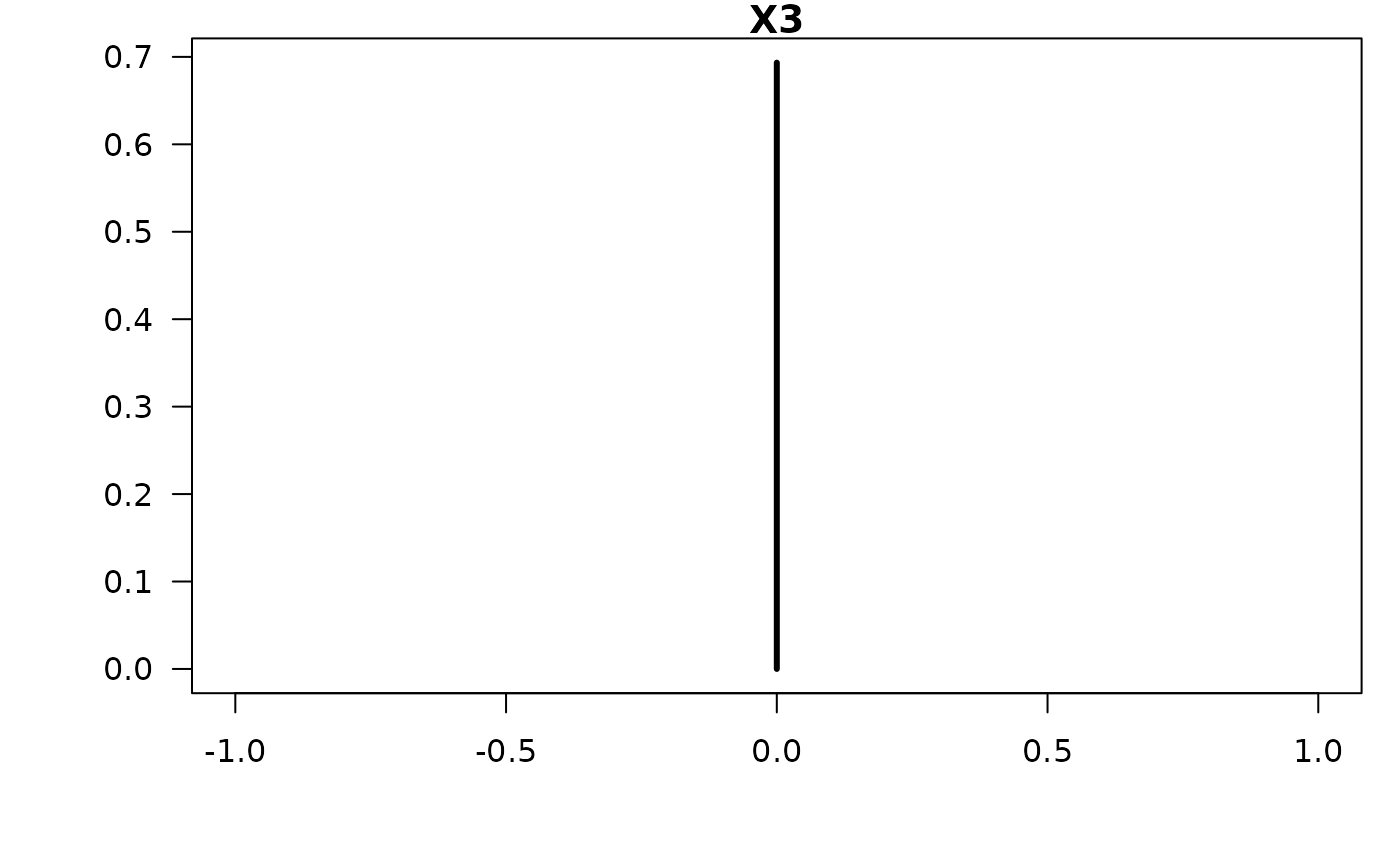
 confint(coef.hald.gprior)
#> 2.5% 97.5% beta
#> Intercept 93.9689432 96.877211 95.4230769
#> X1 1.1922938 1.718554 1.4554239
#> X2 0.5569707 0.755910 0.6564404
#> X3 0.0000000 0.000000 0.0000000
#> X4 0.0000000 0.000000 0.0000000
#> attr(,"Probability")
#> [1] 0.95
#> attr(,"class")
#> [1] "confint.bas"
# To add estimation under Best Predictive Model
confint(coef.hald.gprior)
#> 2.5% 97.5% beta
#> Intercept 93.9689432 96.877211 95.4230769
#> X1 1.1922938 1.718554 1.4554239
#> X2 0.5569707 0.755910 0.6564404
#> X3 0.0000000 0.000000 0.0000000
#> X4 0.0000000 0.000000 0.0000000
#> attr(,"Probability")
#> [1] 0.95
#> attr(,"class")
#> [1] "confint.bas"
# To add estimation under Best Predictive Model